Page 121 - Molecular features of low-grade developmental brain tumours
P. 121
THE CODING AND NON-CODING TRANSCRIPTIONAL LANDSCAPE OF SEGA
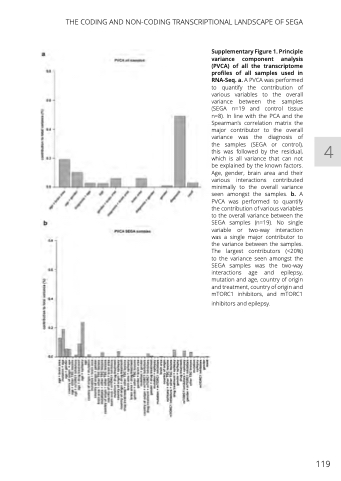
Supplementary Figure 1. Principle variance component analysis (PVCA) of all the transcriptome profiles of all samples used in RNA-Seq. a. A PVCA was performed to quantify the contribution of various variables to the overall variance between the samples (SEGA n=19 and control tissue n=8). In line with the PCA and the Spearman’s correlation matrix the major contributor to the overall variance was the diagnosis of the samples (SEGA or control), this was followed by the residual, which is all variance that can not be explained by the known factors. Age, gender, brain area and their various interactions contributed minimally to the overall variance seen amongst the samples. b. A PVCA was performed to quantify the contribution of various variables to the overall variance between the SEGA samples (n=19). No single variable or two-way interaction was a single major contributor to the variance between the samples. The largest contributors (<20%) to the variance seen amongst the SEGA samples was the two-way interactions age and epilepsy, mutation and age, country of origin and treatment, country of origin and mTORC1 inhibitors, and mTORC1
inhibitors and epilepsy.
119
4