Page 184 - Microbial methane cycling in a warming world From biosphere to atmosphere Michiel H in t Zandt
P. 184
Chapter 8. Species-specific responses of enriched thermokarst lake sediments
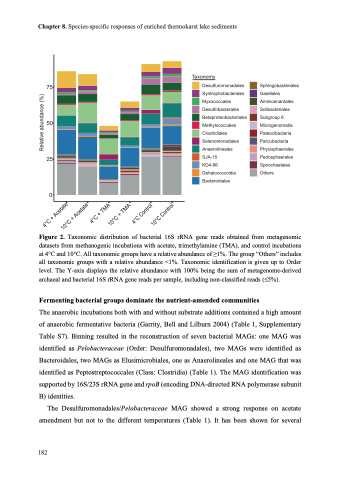
Figure 2. Taxonomic distribution of bacterial 16S rRNA gene reads obtained from metagenomic dFaigta.se2ts from methanogenic incubations with acetate, trimethylamine (TMA), and control incubations at 4°C and 10°C. All taxonomic groups have a relative abundance of ≥1%. The group “Others” includes all taxonomic groups with a relative abundance <1%. Taxonomic identification is given up to Order level. The Y-axis displays the relative abundance with 100% being the sum of metagenome-derived archaeal and bacterial 16S rRNA gene reads per sample, including non-classified reads (£5%).
Fermenting bacterial groups dominate the nutrient-amended communities
The anaerobic incubations both with and without substrate additions contained a high amount of anaerobic fermentative bacteria (Garrity, Bell and Lilburn 2004) (Table 1, Supplementary Table S7). Binning resulted in the reconstruction of seven bacterial MAGs: one MAG was identified as Pelobacteraceae (Order: Desulfuromonadales), two MAGs were identified as Bacteroidales, two MAGs as Elusimicrobiales, one as Anaerolineales and one MAG that was identified as Peptostreptococcales (Class: Clostridia) (Table 1). The MAG identification was supported by 16S/23S rRNA gene and rpoB (encoding DNA-directed RNA polymerase subunit B) identities.
The Desulfuromonadales/Pelobacteraceae MAG showed a strong response on acetate amendment but not to the different temperatures (Table 1). It has been shown for several
182