Page 182 - Microbial methane cycling in a warming world From biosphere to atmosphere Michiel H in t Zandt
P. 182
Chapter 8. Species-specific responses of enriched thermokarst lake sediments
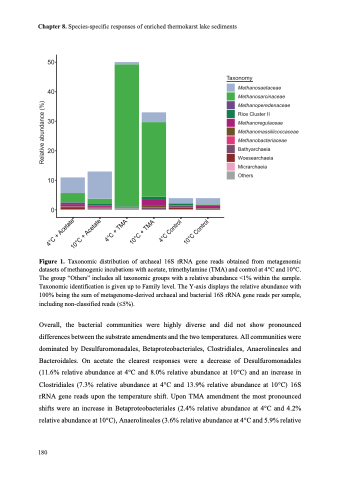
Figure 1. Taxonomic distribution of archaeal 16S rRNA gene reads obtained from metagenomic datasets of methanogenic incubations with acetate, trimethylamine (TMA) and control at 4°C and 10°C.
Fig. 1
The group “Others” includes all taxonomic groups with a relative abundance <1% within the sample. Taxonomic identification is given up to Family level. The Y-axis displays the relative abundance with 100% being the sum of metagenome-derived archaeal and bacterial 16S rRNA gene reads per sample, including non-classified reads (£5%).
Overall, the bacterial communities were highly diverse and did not show pronounced differences between the substrate amendments and the two temperatures. All communities were dominated by Desulfuromonadales, Betaproteobacteriales, Clostridiales, Anaerolineales and Bacteroidales. On acetate the clearest responses were a decrease of Desulfuromonadales (11.6% relative abundance at 4°C and 8.0% relative abundance at 10°C) and an increase in Clostridiales (7.3% relative abundance at 4°C and 13.9% relative abundance at 10°C) 16S rRNA gene reads upon the temperature shift. Upon TMA amendment the most pronounced shifts were an increase in Betaproteobacteriales (2.4% relative abundance at 4°C and 4.2% relative abundance at 10°C), Anaerolineales (3.6% relative abundance at 4°C and 5.9% relative
180