Page 121 - Personalised medicine of fluoropyrimidines using DPYD pharmacogenetics Carin Lunenburg
P. 121
Supplement
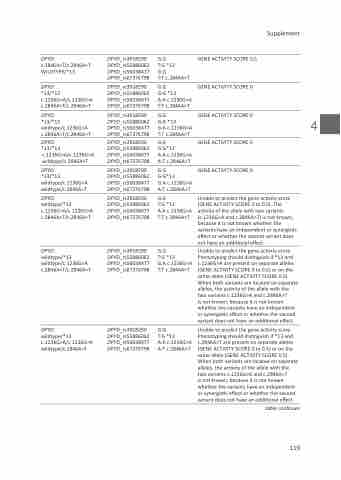
DPYD: c.2846A>T/c.2846A>T WILDTYPE/*13
DPYD:
*13/*13 c.1236G>A/c.1236G>A c.2846A>T/c.2846A>T
DPYD:
*13/*13 wildtype/c.1236G>A c.2846A>T/c.2846A>T
DPYD: *13/*13
c.1236G>A/c.1236G>A wildtype/c.2846A>T
DPYD:
*13/*13 wildtype/c.1236G>A wildtype/c.2846A>T
DPYD:
wildtype/*13 c.1236G>A/c.1236G>A c.2846A>T/c.2846A>T
DPYD:
wildtype/*13 wildtype/c.1236G>A c.2846A>T/c.2846A>T
DPYD:
wildtype/*13 c.1236G>A/c.1236G>A wildtype/c.2846A>T
DPYD_rs3918290 DPYD_rs55886062 DPYD_rs56038477 DPYD_rs67376798
DPYD_rs3918290 DPYD_rs55886062 DPYD_rs56038477 DPYD_rs67376798
DPYD_rs3918290 DPYD_rs55886062 DPYD_rs56038477 DPYD_rs67376798
DPYD_rs3918290 DPYD_rs55886062 DPYD_rs56038477 DPYD_rs67376798
DPYD_rs3918290 DPYD_rs55886062 DPYD_rs56038477 DPYD_rs67376798
DPYD_rs3918290 DPYD_rs55886062 DPYD_rs56038477 DPYD_rs67376798
DPYD_rs3918290 DPYD_rs55886062 DPYD_rs56038477 DPYD_rs67376798
DPYD_rs3918290 DPYD_rs55886062 DPYD_rs56038477 DPYD_rs67376798
G:G
T:G *13
G:G
T:T c.2846A>T
G:G
G:G *13
A:A c.1236G>A T:T c.2846A>T
G:G
G:G *13
G:A c.1236G>A T:T c.2846A>T
G:G
G:G*13
A:A c.1236G>A A:T c.2846A>T
G:G
G:G*13
G:A c.1236G>A A:T c.2846A>T
G:G
T:G *13
A:A c.1236G>A T:T c.2846A>T
G:G
T:G *13
G:A c.1236G>A T:T c.2846A>T
G:G
T:G *13
A:A c.1236G>A A:T c.2846A>T
GENE ACTIVITY SCORE 0,5
GENE ACTIVITY SCORE 0
GENE ACTIVITY SCORE 0
GENE ACTIVITY SCORE 0
GENE ACTIVITY SCORE 0
Unable to predict the gene activity score (GENE ACTIVITY SCORE 0 to 0.5). The activity of the allele with two variants (c.1236G>A and c.2846A>T) is not known, because it is not known whether the variants have an independent or synergistic effect or whether the second variant does not have an additional effect.
Unable to predict the gene activity score. Phenotyping should distinguish if *13 and c.1236G>A are present on separate alleles (GENE ACTIVITY SCORE 0 to 0.5) or on the same allele (GENE ACTIVITY SCORE 0.5). When both variants are located on separate alleles, the activity of the allele with the two variants c.1236G>A and c.2846A>T
is not known, because it is not known whether the variants have an independent or synergistic effect or whether the second variant does not have an additional effect.
Unable to predict the gene activity score. Phenotyping should distinguish if *13 and c.2846A>T are present on separate alleles (GENE ACTIVITY SCORE 0 to 0.5) or on the same allele (GENE ACTIVITY SCORE 0.5). When both variants are located on separate alleles, the activity of the allele with the two variants c.1236G>A and c.2846A>T
is not known, because it is not known whether the variants have an independent or synergistic effect or whether the second variant does not have an additional effect.
table continues
4
119