Page 132 - Molecular features of low-grade developmental brain tumours
P. 132
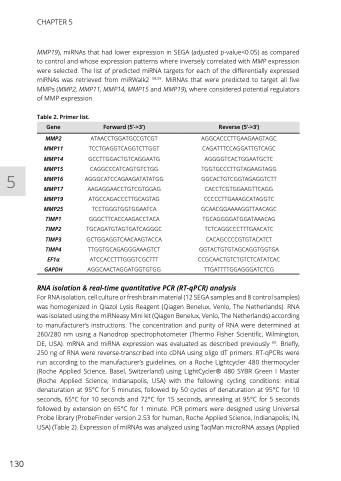
Gene
Forward (5’->3’)
Reverse (5’->3’)
5
130
CHAPTER 5
MMP19), miRNAs that had lower expression in SEGA (adjusted p-value<0.05) as compared to control and whose expression patterns where inversely correlated with MMP expression were selected. The list of predicted miRNA targets for each of the differentially expressed miRNAs was retrieved from miRWalk2 58,59. MiRNAs that were predicted to target all five MMPs (MMP2, MMP11, MMP14, MMP15 and MMP19), where considered potential regulators of MMP expression.
Table 2. Primer list.
MMP2 MMP11 MMP14 MMP15 MMP16 MMP17 MMP19 MMP25 TIMP1 TIMP2 TIMP3 TIMP4 EF1α GAPDH
ATAACCTGGATGCCGTCGT TCCTGAGGTCAGGTCTTGGT GCCTTGGACTGTCAGGAATG CAGGCCCATCAGTGTCTGG AGGGCATCCAGAAGATATATGG AAGAGGAACCTGTCGTGGAG ATGCCAGACCCTTGCAGTAG TCCTGGGTGGTGGAATCA GGGCTTCACCAAGACCTACA TGCAGATGTAGTGATCAGGGC GCTGGAGGTCAACAAGTACCA TTGGTGCAGAGGGAAAGTCT ATCCACCTTTGGGTCGCTTT AGGCAACTAGGATGGTGTGG
AGGCACCCTTGAAGAAGTAGC CAGATTTCCAGGATTGTCAGC AGGGGTCACTGGAATGCTC TGGTGCCCTTGTAGAAGTAGG GGCACTGTCGGTAGAGGTCTT CACCTCGTGGAAGTTCAGG CCCCCTTGAAAGCATAGGTC GCAACGGAAAAGGTTAACAGC TGCAGGGGATGGATAAACAG TCTCAGGCCCTTTGAACATC CACAGCCCCGTGTACATCT GGTACTGTGTAGCAGGTGGTGA CCGCAACTGTCTGTCTCATATCAC TTGATTTTGGAGGGATCTCG
RNA isolation & real-time quantitative PCR (RT-qPCR) analysis
For RNA isolation, cell culture or fresh brain material (12 SEGA samples and 8 control samples) was homogenized in Qiazol Lysis Reagent (Qiagen Benelux, Venlo, The Netherlands). RNA was isolated using the miRNeasy Mini kit (Qiagen Benelux, Venlo, The Netherlands) according to manufacturer’s instructions. The concentration and purity of RNA were determined at 260/280 nm using a Nanodrop spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA). mRNA and miRNA expression was evaluated as described previously 60. Briefly, 250 ng of RNA were reverse-transcribed into cDNA using oligo dT primers. RT-qPCRs were run according to the manufacturer’s guidelines, on a Roche Lightcycler 480 thermocycler (Roche Applied Science, Basel, Switzerland) using LightCycler® 480 SYBR Green I Master (Roche Applied Science, Indianapolis, USA) with the following cycling conditions: initial denaturation at 95°C for 5 minutes, followed by 50 cycles of denaturation at 95°C for 10 seconds, 65°C for 10 seconds and 72°C for 15 seconds, annealing at 95°C for 5 seconds followed by extension on 65°C for 1 minute. PCR primers were designed using Universal Probe library (ProbeFinder version 2.53 for human, Roche Applied Science, Indianapolis, IN, USA) (Table 2). Expression of miRNAs was analyzed using TaqMan microRNA assays (Applied