Page 308 - Personalised medicine of fluoropyrimidines using DPYD pharmacogenetics Carin Lunenburg
P. 308
Chapter 12
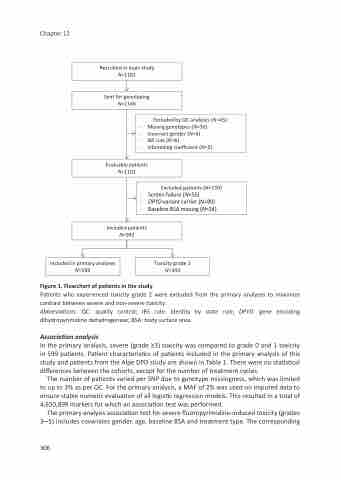
Recruited in main study N=1181
Sent for genotyping N=1146
Excluded by QC analyses (N=45):
- Missing genotypes (N=30)
- Incorrect gender (N=4)
- IBS rule (N=6)
- Inbreeding coefficient (N=5)
Evaluable patients N=1101
Excluded patients (N=159)
- Screen failure (N=55)
- DPYD variant carrier (N=80)
- Baseline BSA missing (N=24)
Included patients N=942
Included in primary analyses N=599
Toxicity grade 2 N=343
Figure 1. Flowchart of patients in the study
Patients who experienced toxicity grade 2 were excluded from the primary analyses to maximize contrast between severe and non-severe toxicity.
Abbreviations: QC: quality control; IBS rule: identity by state rule; DPYD: gene encoding dihydropyrimidine dehydrogenase; BSA: body surface area.
Association analysis
In the primary analysis, severe (grade ≥3) toxicity was compared to grade 0 and 1 toxicity in 599 patients. Patient characteristics of patients included in the primary analysis of this study and patients from the Alpe DPD study are shown in Table 1. There were no statistical differences between the cohorts, except for the number of treatment cycles.
The number of patients varied per SNP due to genotype missingness, which was limited to up to 3% as per QC. For the primary analysis, a MAF of 2% was used on imputed data to ensure stable numeric evaluation of all logistic regression models. This resulted in a total of 4,650,899 markers for which an association test was performed.
The primary analysis association test for severe fluoropyrimidine-induced toxicity (grades 3─5) includes covariates gender, age, baseline BSA and treatment type. The corresponding
306