Page 114 - Personalised medicine of fluoropyrimidines using DPYD pharmacogenetics Carin Lunenburg
P. 114
Chapter 4
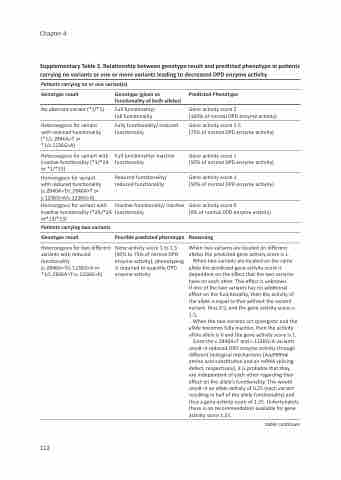
Supplementary Table 3. Relationship between genotype result and predicted phenotype in patients carrying no variants or one or more variants leading to decreased DPD enzyme activity
Patients carrying no or one variant(s)
Genotype result
No aberrant variant (*1/*1)
Heterozygous for variant with reduced functionality (*1/c.2846A>T or *1/c.1236G>A)
Heterozygous for variant with inactive functionality (*1/*2A or *1/*13)
Homozygous for variant with reduced functionality (c.2846A>T/c.2846A>T or c.1236G>A/c.1236G>A)
Homozygous for variant with inactive functionality (*2A/*2A or*13/*13)
Patients carrying two variants
Genotype result
Heterozygous for two different variants with reduced functionality (c.2846A>T/c.1236G>A or *1/c.2846A>T+c.1236G>A)
Genotype (given as functionality of both alleles)
Full functionality/ full functionality
Fully functionality/ reduced functionality
Full functionality/ inactive functionality
Reduced functionality/ reduced functionality
Inactive functionality/ inactive functionality
Possible predicted phenotype
Gene activity score 1 to 1.5 (50% to 75% of normal DPD enzyme activity), phenotyping is required to quantify DPD enzyme activity
Predicted Phenotype
Gene activity score 2
(100% of normal DPD enzyme activity)
Gene activity score 1.5
(75% of normal DPD enzyme activity)
Gene activity score 1
(50% of normal DPD enzyme activity)
Gene activity score 1
(50% of normal DPD enzyme activity)
Gene activity score 0
(0% of normal DPD enzyme activity)
Reasoning
When two variants are located on different alleles the predicted gene activity score is 1.
When two variants are located on the same allele the predicted gene activity score is dependent on the effect that the two variants have on each other. This effect is unknown.
If one of the two variants has no additional effect on the functionality, then the activity of the allele is equal to that without the second variant, thus 0.5, and the gene activity score is 1.5.
When the two variants act synergistic and the allele becomes fully inactive, then the activity ofthe allele is 0 and the gene activity score is 1.
Since the c.2846A>T and c.1236G>A variants result in reduced DPD enzyme activity through different biological mechanisms (Asp949Val amino acid substitution and an mRNA splicing- defect, respectively), it is probable that they
are independent of each other regarding their effect on the allele’s functionality. This would result in an allele activity of 0.25 (each variant resulting in half of the allele functionality) and thus a gene activity score of 1.25. Unfortunately there is no recommendation available for gene activity score 1.25.
table continues
112